We think of DNA as the vital molecules that carry genetic instructions for most living things, including ourselves. But not all DNA encodes proteins; now, we find more and more functions involving non-coding DNA that scientists used to consider “junk.”
A new study suggests that satellite DNA (a type of non-coding DNA arranged in long, repetitive, seemingly meaningless strands of genetic material) may be the reason why different species cannot reproduce successfully.
It seems that the DNA of satellites plays an essential role in keeping all the chromosomes of a cell together in a single nucleus, through the work of cellular proteins.
According to biologists Madhav Jagannathan and Yukiko Yamashita, authors of the new study, this important role is managed differently in each species, leading to genetic incompatibility. The clash between different species-to-species strategies may be what causes chromosomes to disperse outside the nucleus, at least in part, preventing reproduction.
“We propose a unifying framework that explains how the widely observed satellite DNA divergence between closely related species can cause reproductive isolation,” they write in their paper.
This “satellite DNA divergence” has been well established in previous research, prompting suspicions about its role in speciation. In the case of the chimpanzee genome and the human genome, for example, the DNA encoding the proteins is almost identical, while the “junk” DNA is almost completely different.
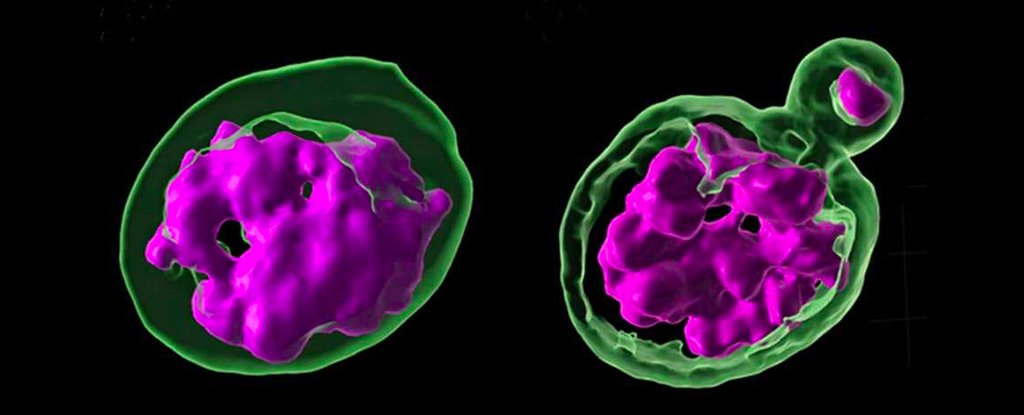
In this new study, we experiment on the fruit fly Drosophila melanogaster, the researchers noted that the removal of a gene that produces a protein called Prod – which binds to a specific fragment of satellite DNA – killed the flies as their chromosomes spread out. of the cell nucleus. However, this crucial piece of satellite DNA is missing from the closest relatives of the flies, who survive well without it.
This suggests that these important non-coding sequences of DNA material have evolved differently between species. To delve deeper, the team examined the family’s hybrid offspring D. melanogaster a female and a male of the close family D. simulations spices.
Flies raised in this way usually die very quickly or end up sterile. In this case, an examination of the tissue of the hybrid offspring confirmed what the researchers suspected: that the chromosomes (the DNA packets needed for reproduction) were also being altered here.
“When we examined those hybrid tissues, it was very clear that their phenotype was exactly the same as if you had altered the satellite chromosomal organization of a pure DNA-mediated species,” says Yamashita, who works at the Massachusetts Institute. of Technology. (MIT).
“The chromosomes were scattered and not encapsulated in a single nucleus.”
Fleeing even further, the researchers produced healthy hybrid flies by removing genes known to damage hybrid offspring (called “hybrid incompatibility genes”) from their parent flies. These incompatibility genes are known to be located in the satellite DNA of the pure species.
Satellite DNA mutates fairly regularly and researchers think that proteins that bind to satellite DNA to hold chromosomes together must evolve to stay up to date. This gives each species its own different strategy when it comes to satellite DNA operations.
The team then wants to try to design a protein that successfully binds to the satellite DNA of two species, keeping the chromosomes where they should be. This could allow viable offspring among these species, but it will take years to do so.
“Our study lays the groundwork for understanding hybrid incompatibility at the cellular level in Drosophila as well as other eukaryotes, ”the researchers write.
The research has been published in Molecular biology and evolution.